Plankton and Particles Flow Cytometry
The guidelines presented below will assist the user in submitting flow cytometry data to SeaBASS. The format described herein allows for Level 2 submissions of flow cytometry data, meaning sums of total abundances of each phytoplankton or bacterial group. Specific metadata headers and field names have been developed for these data. Additionally, detailed instructions are given for the submission of documentation (protocol document, checklist, etc.), images, and additional ancillary files. Greater detail regarding the development and application of these guidelines and requirements can be found in Neeley et al., 2023.
Neeley, A.R., Soto-Ramos, I. and Proctor, C. (2023) Standards and Best Practices for Reporting Flow Cytometry Observations: a technical manual, Version 1.1. Greenbelt, MD., NASA Goddard Space Flight Center, 31pp. DOI: http://dx.doi.org/10.25607/OBP-1864.2.
Table of Contents
Required headers, and fields: FCM
Conditionally required metadata headers
Required metadata headers:
- /associated_archives: List of filenames for all external raw data (e.g., a bundle of .fcs files) and/or scientific files (e.g., a bundle of gate plot images as individual files or all in one large pdf/image file.) Please split files that exceed 5Gb.
Example: /associated_archives=EXPERIMENT_CRUISE_FCM_fcs_associated.tgz,EXPERIMENT_CRUISE_gatePlots_associated.tgz - /associated_archive_types: Provide a value or list of text terms to describe the contents of each associated_archive file. Commonly used types are 'raw' for raw (.fcs) files, 'ancillary_media' for images, or 'metadata' for other supporting files or code.
Example: /associated_archive_types=raw,ancillary_media
Required "if applicable" metadata headers:
- /associated_files: The value is the name of the specific source file used for the scientific analysis. Include this header if all the data in this file came from the same sample or .fcs file. However, if multiple samples were used, then skip this header and instead use the equivalent field associated_files so you can name the specific file on each data row.
- /associated_file_types: This header must be used in conjunction with associated_files. The entry here should describe the data type within the associated_files. For the purposes of .fcs files, the format would be ‘raw’, images would be ‘ancillary_media’ and any other supportive information would go under ‘metadata’.
- /eventID: a unique identifier associated with the sample as an event.
Conditionally required data fields
This section describes how to format the data table for flow cytometry observations. An external, machine-readable, and resolvable identifier, or object number, that returns nomenclatural (not taxonomic) details of a name (scientificNameID) should also be included for each organism. Moreover, the field names measurementValue and measurementValueID, which provide morphological and functional information, were added to remain interoperable with the Ocean Biodiversity Information System (OBIS) and align with Darwin Core terminology when possible (See subchapter 3.2 in Neeley et al. 2023). The complete and updated lists of NERC vocabulary can be found here. The date that the vocabulary was accessed and a link to the most current NERC vocabulary must be included in the checklist and in the comments section of the data file.
Required fields:
- volume_analyzed_ul (ul): Volume that was processed through the flow cytometer in units of microliters (ul). This field name can also be used as a header when a single sample is included per submission.
- data_provider_category_manual (none): A category used by the data provider to name the organism for a manual identification, not necessarily a scientific name.
- scientificName_manual (none): A scientific name from a recognized taxonomic reference database (e.g., World Register of Marine Species, AlgaeBase) at the lowest level that matches the data provider's category, for a manual identification matched to scientificNameID.
- scientificNameID_manual (none): A Life Science Identifier (LSID) from a recognized taxonomic reference database (e.g., World Register of Marine Species, AlgaeBase) at the lowest level that matches the data provider's category for manual identification.
- abun (cells/L): Concentration of cells observed in units of cells per liter.
- FCM_total_events_L (events/L): Total events captured by the instrument. This value should not include any data filtering/removal
- FCM_cell_percentage (%): The total amount of cells from all gates divided by the total amount of cells or events
- Elapsed_time (s): The amount of time the instrument processes a sample. This will be after any clipping of time to account for inconsistent flow of the instrument
Recommended but optional fields:
- measurementValue (none): Vocabulary that describes a cluster of cells by morphological characteristics (e.g., size and fluorescence) rather than by taxonomy and is recognized by the Natural Environment Research Council (NERC). The alternative label must be used here.
- measurementValueID (none) The URI associated with the NERC vocabulary is defined in measurementValue.
- data_provider_category_manual: (none) A category used by the data provider to name the organism for a manual identification, not necessarily a scientific name.
Required "if applicable" fields:
- /associated_files: The value is the name of the specific source file (.fcs) used for the scientific analysis.
- /associated_file_types: This header must be used in conjunction with associated_files. The entry here should describe the data type within the associated_files. For the purposes of .fcs files, the format would be ‘raw’, images would be ‘ancillary_media’ and any other supportive information would go under ‘metadata’.
Required extra documents, FCM
Submission checklist:
- The data file must include a data table and appropriate metadata headers and field names
- Protocol document
- Data set checklist
- Ancillary data files (raw files (.fcs), 1 required gate plot and potential additional gate plots, etc.)
- Text file containing list of assessed IDs for manual classification
Protocol and checklist documents
Submission of raw files and additional metadata
FCM submission will be required to include:
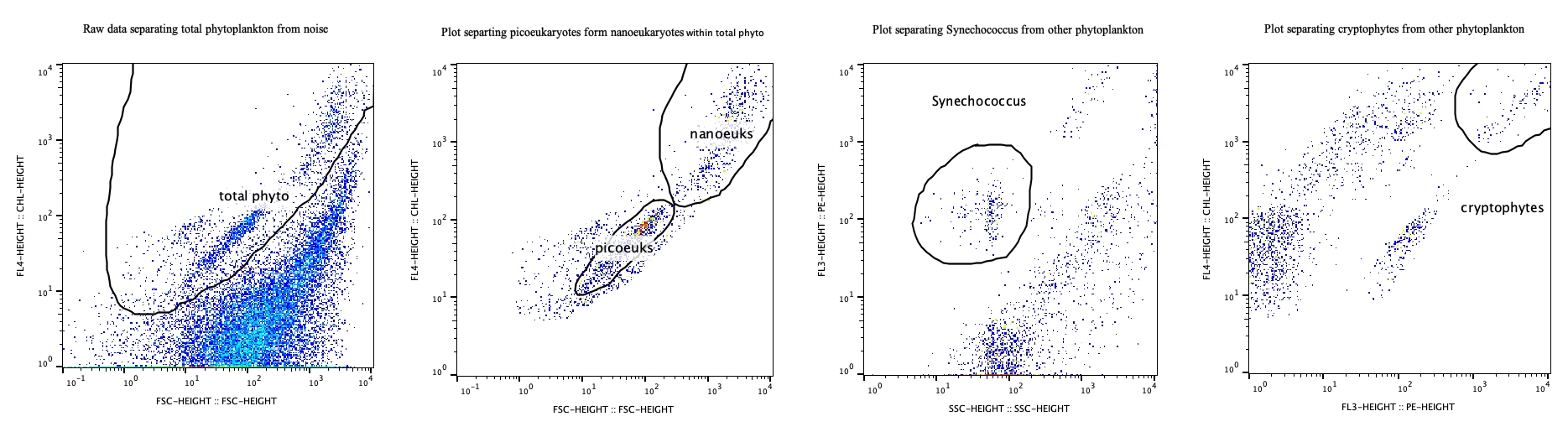
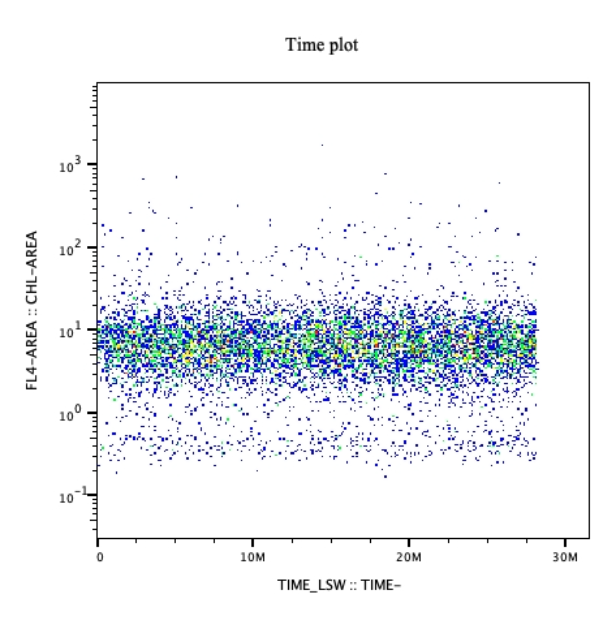
At least 1 scatter type plot with the axes being any channels and scatter type (FSC, SSC) used to create a gate (region created by the analyst to calculate cells/events per mL). The other plot required will have time as the x-axis and the analyst’s preferred channel as the y axis (typically chl-a or similar for non-stained samples).
- Plot showing all gates used for analysis (share multiple plots if multiple axes were used)
- gate_plot can be used for the plot(s) that show where gates are positioned by the analyst. Only one plot per channel selection is required for the entirety of the cruise, showing how the gates show up against the raw data.
- These plots should include any gate created by the analyst, even if they are not used for every sample.

- We recommend naming this file cruiseName_timeplot, for a scatter plot that has time as the X axis and the analysts preferred channel on the Y axis.
- One plot per cruise is strongly recommended with additional plots per/sample optional as well.
- Please note that the x- axis of this plot type should be in seconds if possible.
- Sample_plot_XXXX_sampleID can be used to name plots that show the FCM data within the gates provided by the analyst if . The XXXX would be the name of the laser type used to calculate the cells/mL of that gate.
- An example would be sample_plot_red_4002. One plot per channel type per cr will be submitted with the data. Other channel types can be orange, blue, UV, chl-a, or the wavelength of the laser chosen.
- One plot per sample showing events over time (no gates)
Assessed IDs list
The following table is an example of an assessed ID table submission for flow cytometry observations.
|
measurementValue |
measurmentValueID |
data_provider_category |
scientificName_manual |
scientificNameID_manual |
|
Red Pico |
http://vocab.nerc.ac.uk/collection/F02/current/F0200004/ |
picoeukaryote |
Eukaryota |
urn:lsid:algaebase.org:taxname:86701 |
|
OraPicoProk |
http://vocab.nerc.ac.uk/collection/F02/current/F0200003/ |
Synechococcus |
Synechococcus |
urn:lsid:marinespecies.org:taxname:160572 |
|
OraNano |
http://vocab.nerc.ac.uk/collection/F02/current/F0200006/ |
Cryptophyceae |
Cryptophyceae |
urn:lsid:marinespecies.org:taxname:17639 |
|
HetHNA |
http://vocab.nerc.ac.uk/collection/F02/current/F0200010/ |
heterotrophic_prokaryote_HNA |
Prokaryota |
urn:lsid:algaebase.org:taxname:86700 |